exp(-Ea/k_B T), lowering the barrier by one electron volt (23 kcal/mol) has a dramatic effect.
But, how is this lowering of the transition state achieved? There is no doubt that simple electrostatic effects can make a major contribution, as emphasised by Ariel Warshel. From the point of view of quantum physics, this is rather "boring". But, thats good science: going with the best and simplest explanation.
However, that is not the whole story, and particularly not for all enzymes.
One enzyme that is attracting significant interest is (KSI) keto-steroid isomerase, highlighted by nice work from the groups of Steve Boxer and Daniel Herschlag at Stanford. This enzyme catalyses the conversion of cholesterol into steroids such as oestrogen and testosterone.
Understanding has advanced by clever studies performing mutations [replacing specific amino acids in the protein by others] and replacing the reactant with a simpler phenol group with tuneable acidity, and seeing how the enzyme structure and catalytic power change.
Here are two short commentaries that put that work in context.
What Governs Enzyme Activity? For One Enzyme, Charge Contributes Only Weakly
Richard Robinson
Biochemistry: Enzymes under the nanoscope
Anthony J. Kirby and Florian Hollfelder
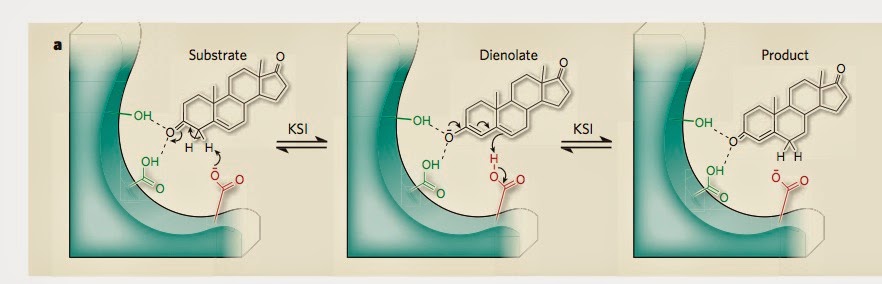
The figure below [taken from the second commentary] how the substrate [reactant] is hydrogen bonded to two tyrosine [amino acids] molecules. The hydrogen network is crucial to the stabilisation of the transition state (middle panel).
That commentary emphasises how small changes in the hydrogen bond lengths [as little as 0.1 Angstroms] produce significant changes in the enzyme catalytic power. But, even engineering such a
small decrease is difficult.
This result has wide-reaching implications: it defines experimentally the distance scale on which enzymes can distinguish geometric rearrangements of atoms, and determines the energetic consequences of this constraint. The picometre-precision of KSI also explains why protein engineering to produce enzymes that have new or altered functions has proved so difficult.
The last statement refers to the grand challenge of trying to do "bio-mimetics" and produce artificial catalysts that could have the incredible power of enzymes.
This structural sensitivity also prevents a significant challenge to structural biology [a.ka. protein crystallography] and computational molecular modelling. Distance resolutions of 0.1 A are at the boundary of the best protein X-ray crystallography. For hydrogen bonds density functional theory based calculations are not that reliable on this length scale. Classical molecular dynamics cannot even describe moderate to short bonds.
I am particularly interested in KSI because of the presence of short to medium strength hydrogen bonds in which quantum nuclear effects play a role. The figure below is taken from this paper.
For such bonds changes in H-bond lengths of 0.1A can produce significant changes in the proton transfer potential.
Furthermore, the interpretation of hydrogen-deuterium isotope substitution experiments will be complicated by the secondary geometric isotope effect.


No comments:
Post a Comment